PROphet® is a unique plasma proteomics profiling platform that predicts response to therapy
to guide clinical decision-making related to first-line immunotherapies
to guide clinical decision-making related to first-line immunotherapies
A new dimension to personalization
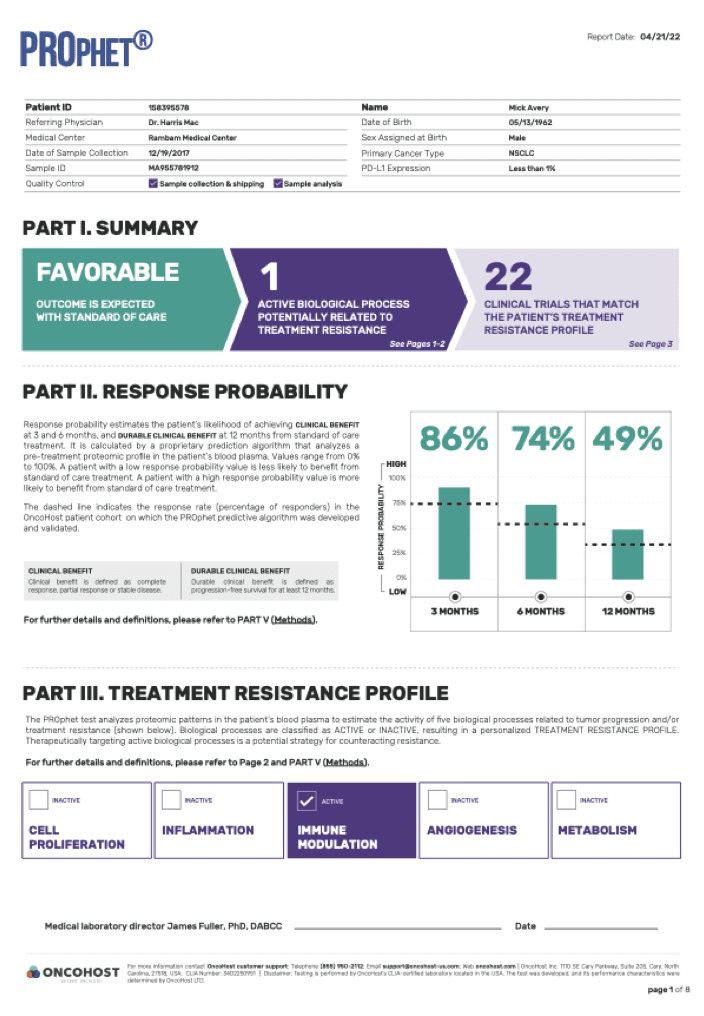
PROphet® analyzes proteomic changes in blood samples to monitor the dynamics of biological processes induced by the patient (i.e., the host) in response to a given cancer therapy. This proteomic profile is highly predictive of individual patient outcomes, thus enabling personalized treatment planning.
PROphet® also identifies potential drug targets, advancing the development of novel therapeutic strategies as well as rationally based combination therapies.
PROphet® also identifies potential drug targets, advancing the development of novel therapeutic strategies as well as rationally based combination therapies.
Currently available for the following indications
Non-Small Cell
Lung Cancer (NSCLC)
Lung Cancer (NSCLC)
Melanoma
(Coming Soon)
(Coming Soon)
With one pre-treatment blood test, PROphet® provides
a report addressing three important clinical questions:
a report addressing three important clinical questions:
01.
Will my patient respond
to their planned treatment?
to their planned treatment?
02.
Why might my patient be
resistant to the recommended
treatment plan and
not respond as desired?
resistant to the recommended
treatment plan and
not respond as desired?
03.
What can I do to mitigate this
patient’s resistance to treatment,
and potentially improve their
outcome?
patient’s resistance to treatment,
and potentially improve their
outcome?
We apply proprietary computational models that analyze the proteomic patterns in the patient’s plasma and identify those proteins that are associated with resistance to treatment. Such proteins participate in different biological processes in the body that can be targeted in order to increase the chances of response.

How it works

Simple pre-treatment
blood test collected
at patient's own home
blood test collected
at patient's own home
High-throughput
proteomic assay
proteomic assay
Bioinformatics and
machine learning
machine learning
The PROphet® report with
response prediction and
actionable clinical insights
is sent to the oncologist
response prediction and
actionable clinical insights
is sent to the oncologist
The blood sample is analyzed and processed at
OncoHost’s CLIA* registered lab in Cary, North Carolina
OncoHost’s CLIA* registered lab in Cary, North Carolina
*CLIA number: 34D2250951
The personalized report has three main sections:
Resistance Biology Mapping
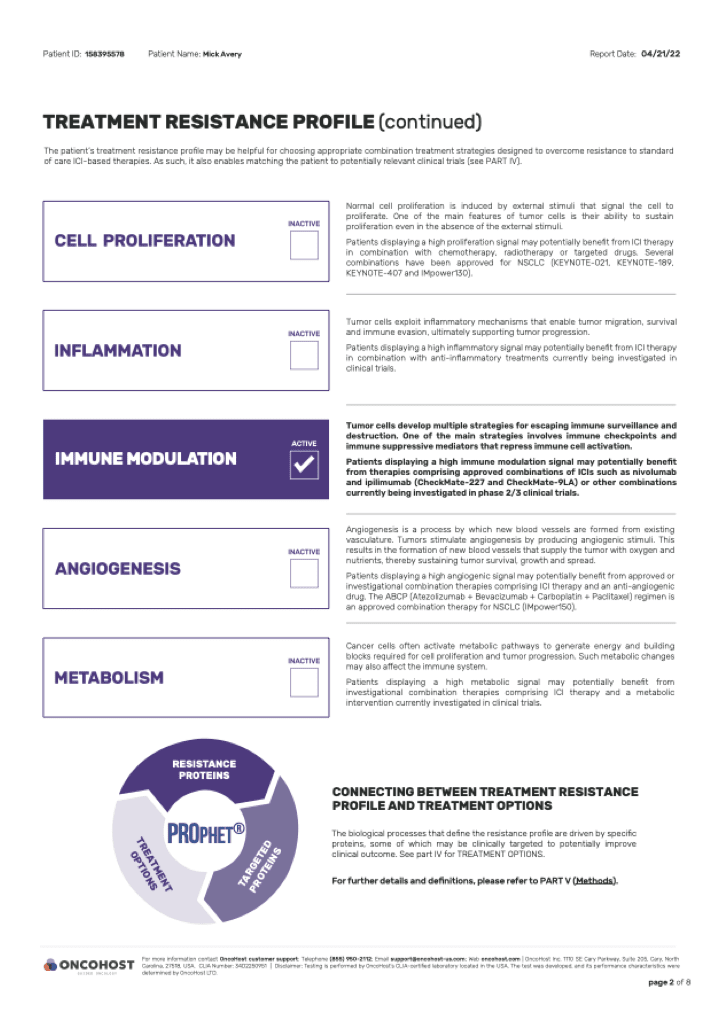
02.
Resistance biology mapping
to provide a clear resistance
mechanism
to provide a clear resistance
mechanism
Actionable Clinical Insights
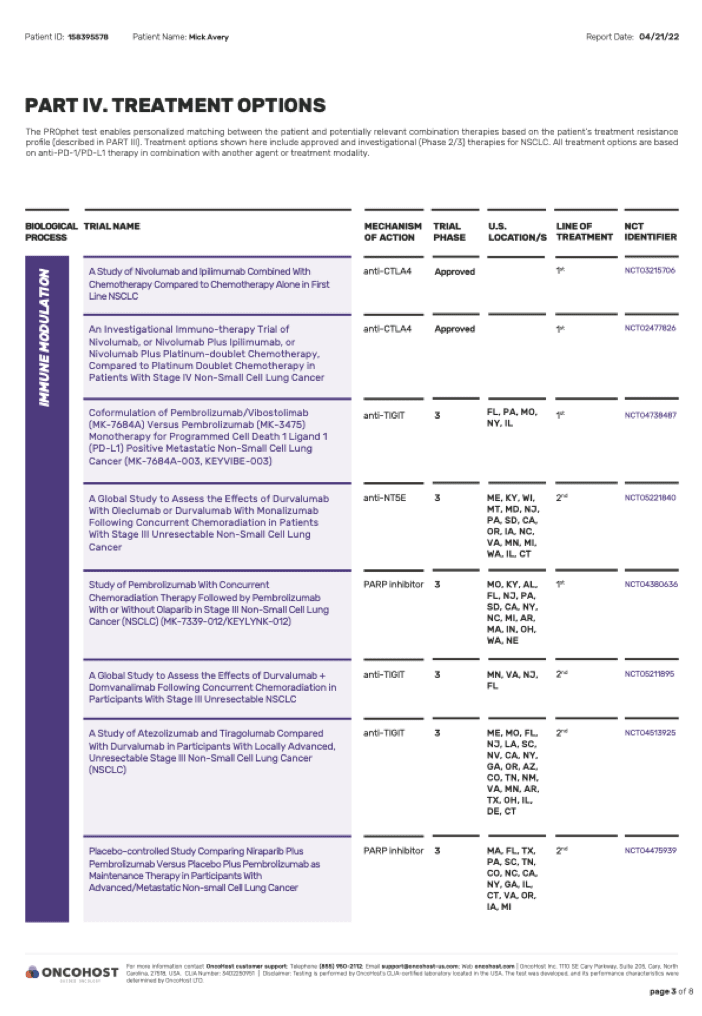
03.
Actionable clinical insights including
approved treatment combinations or
potential relevant clinical trials
approved treatment combinations or
potential relevant clinical trials
Clinical Validation
The PROphet® platform was developed using patient blood samples and clinical data collected within the framework of an ongoing clinical study conducted by OncoHost (PROPHETIC; NCT04056247). In brief, pre-treatment blood samples were collected from 339 advanced stage non-small cell lung cancer (NSCLC) patients undergoing anti-PD-(L)1 immunotherapy.
Patient clinical data were recorded, including demographics, clinical characteristics, and response to treatment. Blood plasma was profiled by the SomaScan® Discovery Assay v4.11-2, and the resulting proteomic profiles were analyzed in conjunction with patient clinical data. Specifically, proprietary AI algorithms were developed to identify proteomic patterns associated with response to anti-PD-(L)1 treatment, as well as to identify potential resistance mechanisms. Algorithm development and validation is described in more detail below.
Patient clinical data were recorded, including demographics, clinical characteristics, and response to treatment. Blood plasma was profiled by the SomaScan® Discovery Assay v4.11-2, and the resulting proteomic profiles were analyzed in conjunction with patient clinical data. Specifically, proprietary AI algorithms were developed to identify proteomic patterns associated with response to anti-PD-(L)1 treatment, as well as to identify potential resistance mechanisms. Algorithm development and validation is described in more detail below.